Abstract
Introduction: We described the mutational spectrum of ATLL in North American patients at ASH 2016 and found that these patients have a heterogenous mutational spectrum (abstract #1758). We also noted some significant differences in comparison to Japanese ATLL cases (Kataoka et al. 2015 PMID: 26437031). In this abstract, we determine the mutational landscape of larger cohort of 27 cases and report on the presence of distinct variants in epigenetic and histone modifying genes.
Methods: Twenty-seven North American primary ATLL patient samples (44% acute, 41% lymphomatous and 15% chronic/smoldering) and 7 Japanese cell lines were analyzed by targeted sequencing of 173 recurrently mutated genes in cancer (Genoptix) at a mean sequencing depth of 500X coverage.
We then carried out proliferation assays with decitabine on 4 Japanese derived cell lines and 5 primary patient samples and belinostat on 1 cell line and 2 primary patient samples. We also did synergy studies with doxorubicin on 3 sensitive samples. R index was calculated for assessing synergy (Romanelli et al. 1998 PMID: 9523734).
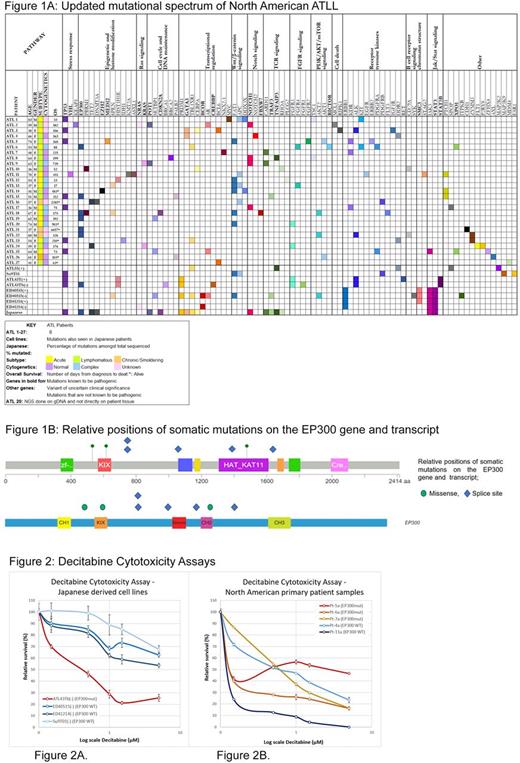
Results: We found a high number of epigenetic and histone modifying gene mutations (EP300, TET2, EZH2, MED12, PBRM1, DNMT3A, KMT2A, HIST1H1E, SPEN, IDH1 and ASXL1) in our patients, 15/27 patients (55.5%). Amongst these genes EP300 had the highest frequency of mutations and was present in 22% of patients. (Figure 1) In comparison, the Japanese cohort of 370 cases had 5.7% cases with EP300 mutations. (OR: 4.1; 95% CI: 1.5099 to 10.9492; p = 0.0055)
The JAK/STAT pathway had significant number of mutations in the Japanese patients (25% had STAT3 mutations and 5% had JAK3 mutations) whereas in our North American cohort there were no STAT3 mutations and 1 patient had a JAK3 mutation (3.7%). The overall percentage of TP53 mutations was similar in both groups 17.8% in the Japanese and 25.9% in the North American patients. The presence of TP53 mutations was associated with a worse overall survival in our patients (p-value: 0.045) and has been described previously (Magalhaes et al. 2015 PMID: 25573202).
EP300 is a histone acetyltransferase that acetylates histones and leads to activation of gene transcription. Inactivating mutations in EP300 can lead to aberrant gene silencing and can be associated with increased DNA methylation. A high prevalence of EP300 mutations prompted us to test the sensitivity of ATLL cell lines and primary samples to DNA methyltransferase inhibitor (Decitabine) and histone deacetylase inhibitor (Belinostat). Among the 4 cell lines tested, only the ATL43Tb(-) cell line with EP300 mutation was sensitive to the growth inhibitory effects of decitabine (IC50 < 1 uM) whereas the IC50 was not reached in resistant samples (three Japanese derived cell lines, ED40515(-) and ED41214(-) and Su9T01(-)) (Figure 2A). Since the cell lines were derived from Japanese ATLL, next, we established long term cultures of primary ATLL cells from North American cases and used them for functional studies. For primary samples, we observed that all samples (n=5) were sensitive to decitabine with IC50 < 1 uM. Furthermore, the three samples (EP300 mutant) were also sensitive to HDAC inhibitor Belinostat with an IC50 of upto 0.5 uM in all samples. Next, we tested the efficacy of combining decitabine with chemotherapy. The combination of doxorubicin and decitabine showed synergy with an R index >1 in three EP300 mutant samples tested Pt-5a, Pt-7a and ATL43Tb(-).
Conclusions: Mutational analysis of the largest cohort of North American ATLL samples shows a distinct variant landscape with a high prevalence of mutations affecting epigenetic modifiers. EP300 is a histone acetyltransferase that is mutated in 22% of samples and is significantly higher in the North American cohort when compared to the Japanese cases.
We also determined that majority of primary ATLL samples and EP300 mutant cell lines tested are sensitive to decitabine. The combination with doxorubicin shows synergy.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.